[Published online Journal of Computer Chemistry, Japan Vol.17, 130-132, by J-STAGE]
<Title:> CD44におけるヒアルロン酸結合機構の解明
<Author(s):> リントゥルオト 正美, 堀岡 洋太, 本郷 紗記, リントゥルオト ユハミカエル
<Corresponding author E-Mill:> masami(at)kpu.ac.jp
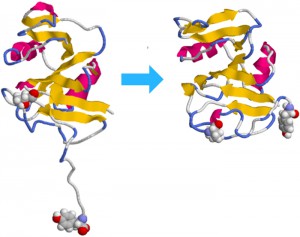
<Abstract:> The hyaluronan (HA) binding mechanism on CD44 was investigated by using molecular dynamics simulation. The Ordered (O) and partially disordered (PD) structures have been reported for CD44 HA binding domain (HABD). Two binding forms were investigated for the HA binding for O and PD structures in this study. O and PD structures show different HA binding structures, and the HA binding affinity on the PD structure was larger than that on O structure. The mobility of HA molecule on CD44 was high, especially for O structure. High affinity of HA binding on PD structure is suggested to regulate the rate of cell rolling under blood flow.
<Keywords:> Keywords CD44, Hyaluronan, Cell rolling, Molecular dynamics, Protein
<URL:> https://www.jstage.jst.go.jp/article/jccj/17/3/17_2018-0027/_article/-char/ja/