[Published online Journal of Computer Chemistry, Japan Vol.17, 228-231, by J-STAGE]
<Title:> 特徴ベクトルから分子構造を生成するアルゴリズム
<Author(s):> 陸 明浩, 安田 耕二
<Corresponding author E-Mill:> yasudak(at)imass.nagoya-u.ac.jp
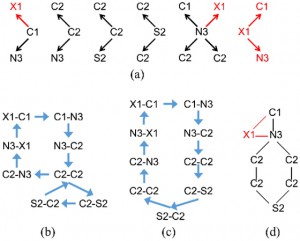
<Abstract:> Machine learning for predicting properties of molecules and designing molecules with desired properties requires suitable numerical representation of molecular structure. In this study we propose to represent a molecular graph as a high dimensional feature vector by counting the number of substructures of a molecule, and report conditions that these vectors should satisfy. The vectors are expanded by the lattice basis extracted from the feature vectors of a large molecular dataset. In addition, an efficient algorithm to reconstruct molecular structures from the feature vector is presented. The latent space provides a continuous and concise representation of molecular graphs, and the representation allows smooth interpolation between two molecules. The proposed method has wide application to studies of quantitative structure activity relationships and machine learning of molecules.
<Keywords:> KEYWORDS Machine learning, Feature vector, Molecular graph reconstruction, De Bruijn graph, Lattice basis
<URL:> https://www.jstage.jst.go.jp/article/jccj/17/5/17_2018-0056/_article/-char/ja/