[Published online Journal of Computer Chemistry, Japan Vol.18, 211-213, by J-STAGE]
<Title:> 分子軌道計算と機械学習によるフェルラ酸の抗酸化作用の研究
<Author(s):> 寺前 裕之, 玄 美燕, 山下 司, 高山 淳, 岡﨑 真理, 坂本 武史
<Corresponding author E-Mill:> teramae(at)gmail.com
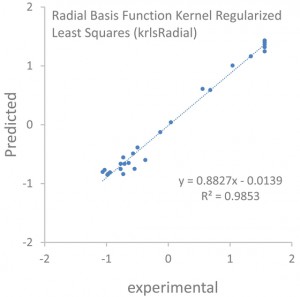
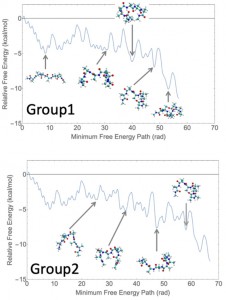
<Abstract:> Ferulic acid is known to have strong antioxidant properties. In the present study, we investigate the electronic structures of ferulic acid and its radical species extracting the hydrogen atom from its phenolic hydroxyl group. The relation of the results by several machine learning models using R/caret package, such as partial least squares, random forest, radial basis function kernel regularized least squares, and baysian regularized neural network, with the radical scavenging activity with the DPPH reagent, IC50, measured by Sakamoto et al. is discussed. We found all four methods gave reasonable correlation coefficients which means the possible prediction of the IC50 values with the results of the molecular orbital calculations only.
<Keywords:> Ferulic acid, Radical scavenging activity, Hamiltonian algorithm, Molecular orbital method, Machine learning, Random forest model
<URL:> https://www.jstage.jst.go.jp/article/jccj/18/5/18_2019-0034/_article/-char/ja/