[Published online Journal of Computer Chemistry, Japan -International Edition Vol.10, -, by J-STAGE]
<Title:> Reconstruction of Four-Body Statistical Pseudopotential for Protein-Peptide Docking
<Author(s):> Tae YAMAMOTO, Yasuhiro IKABATA, Hitoshi GOTO
<Corresponding author E-Mill:> gotoh(at)tut.jp
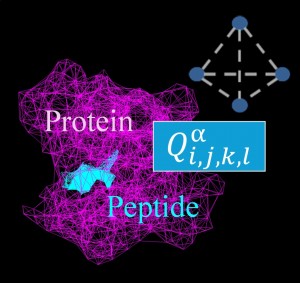
<Abstract:> We constructed the four-body statistical pseudopotential proposed by Krishnamoorthy and Tropsha, which is a type of coarse-grained potential previously used for protein-peptide docking, using different data sets. The first data set is composed of crystal structures of proteins that satisfy the conditions specified using PISCES, a protein sequence culling server. The second data set consists of crystal structures of protein-protein complexes obtained from the PDBbind-CN database. The four-body potential has 44275 patterns of scores, depending on the type of amino acid in the quadruplet of residues and the continuity of amino acid in the sequence of protein or peptide. While the PISCES-based data set covers almost all patterns, docking simulations revealed that both potentials provided comparable accuracy in reproducing experimentally determined peptide binding poses.
<Keywords:> Docking simulation, Protein-peptide complex, Four-body statistical pseudopotential, Coarse-grained potential, Peptide binding pose
<URL:> https://www.jstage.jst.go.jp/article/jccjie/10/0/10_2023-0039/_html
<Title:> Reconstruction of Four-Body Statistical Pseudopotential for Protein-Peptide Docking
<Author(s):> Tae YAMAMOTO, Yasuhiro IKABATA, Hitoshi GOTO
<Corresponding author E-Mill:> gotoh(at)tut.jp
<Abstract:> We constructed the four-body statistical pseudopotential proposed by Krishnamoorthy and Tropsha, which is a type of coarse-grained potential previously used for protein-peptide docking, using different data sets. The first data set is composed of crystal structures of proteins that satisfy the conditions specified using PISCES, a protein sequence culling server. The second data set consists of crystal structures of protein-protein complexes obtained from the PDBbind-CN database. The four-body potential has 44275 patterns of scores, depending on the type of amino acid in the quadruplet of residues and the continuity of amino acid in the sequence of protein or peptide. While the PISCES-based data set covers almost all patterns, docking simulations revealed that both potentials provided comparable accuracy in reproducing experimentally determined peptide binding poses.
<Keywords:> Docking simulation, Protein-peptide complex, Four-body statistical pseudopotential, Coarse-grained potential, Peptide binding pose
<URL:> https://www.jstage.jst.go.jp/article/jccjie/10/0/10_2023-0039/_html